Poster – Evaluation of a Next-Generation Sequencing Metagenomics Assay to Detect and Quantify DNA Viruses in Plasma from Transplant Patients
Enabling NGS-based metagenomics in the clinical virology laboratory
Poster Presentation given at the Clinical Virology Symposium 2019 Learn more about Galileo™Evaluation of a Next-Generation Sequencing Metagenomics Assay to Detect and Quantify DNA Viruses in Plasma from Transplant Patients
This poster was presented at the Clinical Virology Symposium on May 6th, 2019 by Dr. Soya S. Sam, a member of Dr. Angela Caliendo team from the Warren Alpert School of Medicine at Brown University.
Abstract:
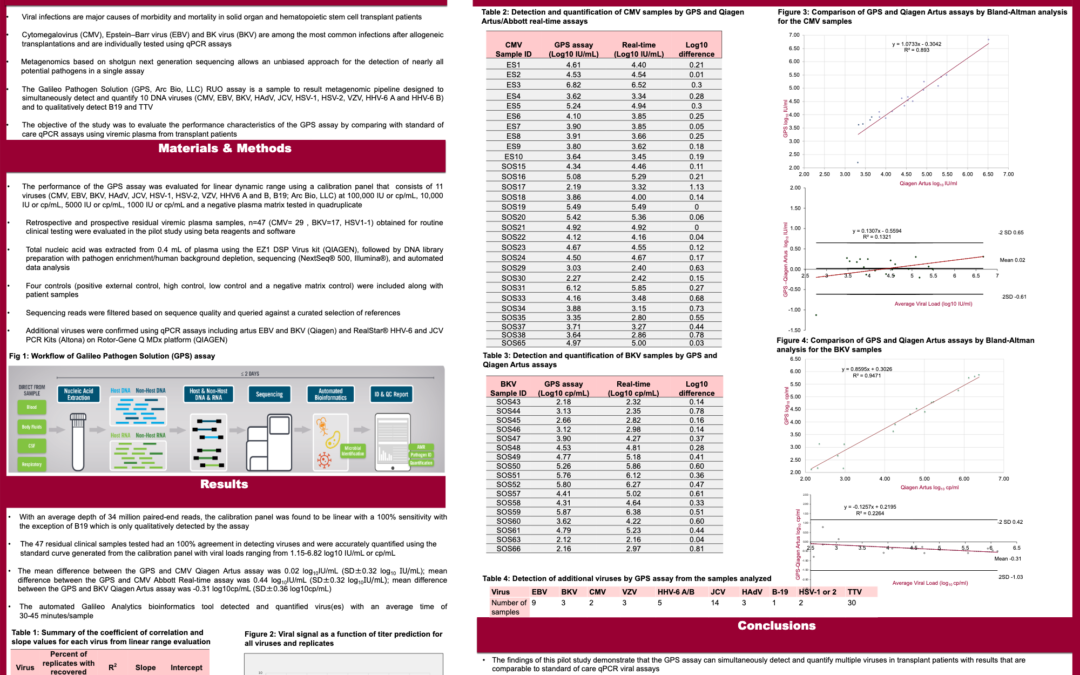
“Viral infections are major causes of morbidity and mortality in solid organ and hematopoietic stem cell transplant patients Cytomegalovirus (CMV), Epstein–Barr virus (EBV) and BK virus (BKV) are among the most common infections after allogeneic transplantations and are individually tested using qPCR assays.
Metagenomics based on shotgun next-generation sequencing allows an unbiased approach for the detection of nearly all potential pathogens in a single assay. The objective of the study was to evaluate the performance characteristics of the Galillo™-Transplant assay by comparing with standard-of-care qPCR assays using viremic plasma from transplant patients.”
In this presentation you will learn:
- What is NGS-based metagenomics (mNGS) is and how it can be applied to pathogen detection
- How mNGS compares with and improves upon qPCR as a detection platform
- What the mNGS workflow looks like when deployed in your laboratory
- Why deploying mNGS into your laboratory makes sense
- How Arc Bio has approached mNGS with its Galileo™ platform